3. Step-by-step analysis (ChIP-seq, human)#
Here we show the step-by-step ChIP-seq analysis using Churros. See also the sample scripts in the tutorial on GitHub.
Note
churros.sif). Please add singularity exec churros.sif before each command below.singularity exec churros.sif download_genomedata.sh3.1. Prepare sample list#
samplelist.txt is a tab-delimited file (TSV) that describes the sample labels and the path to the corresponding fastq files.
HepG2_H2A.Z fastq/SRR227639.fastq.gz
HepG2_H3K4me3 fastq/SRR227563.fastq.gz
HepG2_H3K27ac fastq/SRR227575.fastq.gz
HepG2_H3K27me3 fastq/SRR227598.fastq.gz
HepG2_H3K36me3 fastq/SRR227447.fastq.gz
HepG2_Control fastq/SRR227552.fastq.gz
Churros has a script gen_samplelist.sh to make the initial samplelist.txt.
gen_samplelist.sh fastq/ > samplelist.txt
When using paired-end fastqs, use the second and the third columns to specify the R1 (F3) and R2 (F5) fastqs like this:
HepG2_H2A.Z fastq/SRR227639_1.fastq.gz fastq/SRR227639_2.fastq.gz
HepG2_H3K4me3 fastq/SRR227563_1.fastq.gz fastq/SRR227563_2.fastq.gz
HepG2_H3K27ac fastq/SRR227575_1.fastq.gz fastq/SRR227575_2.fastq.gz
HepG2_H3K27me3 fastq/SRR227598_1.fastq.gz fastq/SRR227598_2.fastq.gz
HepG2_H3K36me3 fastq/SRR227447_1.fastq.gz fastq/SRR227447_2.fastq.gz
HepG2_Control fastq/SRR227552_1.fastq.gz fastq/SRR227552_2.fastq.gz
gen_samplelist.sh -p makes the samplelist.txt for paired-end samples.
gen_samplelist.sh -p fastq/ > samplelist.txt
3.2. Prepare sample pair list#
samplepairlist.txt is a comma-delimited file (CSV) that describes the ChIP/Input pairs as follows:
ChIP-sample label
Input-sample label
prefix
peak mode
HepG2_H2A.Z,HepG2_Control,HepG2_H2A.Z,sharp
HepG2_H3K4me3,HepG2_Control,HepG2_H3K4me3,sharp
HepG2_H3K27ac,HepG2_Control,HepG2_H3K27ac,sharp
HepG2_H3K27me3,HepG2_Control,HepG2_H3K27me3,broad
HepG2_H3K36me3,HepG2_Control,HepG2_H3K36me3,broad
ChIP and input sample labels should be identical to those in samplelist.txt.
prefix is used for the output files.
peak mode is either [sharp|broad|sharp-nomodel|broad-nomodel]. This parameter is used for peak calling by MACS2.
Input samples can be omitted if unavailable.
HepG2_H2A.Z,,HepG2_H2A.Z,sharp
HepG2_H3K4me3,,HepG2_H3K4me3,sharp
HepG2_H3K27ac,,HepG2_H3K27ac,sharp
HepG2_H3K27me3,,HepG2_H3K27me3,broad
HepG2_H3K36me3,,HepG2_H3K36me3,broad
In addition, Churros also has a script gen_samplepairlist.sh to make the initial template of samplepairlist.txt.
gen_samplepairlist.sh samplelist.txt > samplepairlist.txt
3.3. churros_mapping: mapping reads#
churros_mapping takes FASTQ and maps reads to the genome specified by Bowtie2 by default.
The mapped reads are then quality-checked and converted to BigWig files.
build=hg38
Ddir=Referencedata_hg38
# mapping
$sing churros_mapping -p 12 exec samplelist.txt $build $Ddir
# output QC stats
$sing churros_mapping header > churros.QCstats.tsv
$sing churros_mapping stats samplelist.txt $build $Ddir >> churros.QCstats.tsv
- Output
bam/ … map files (BAM format in default) and index files
sspout/ … output of SSP (strand-shift profile) for quality check
bigWig/ … bigWig files (100 bp, 5 kbp and 100 kbp bins by default) with raw count (
RawCount) and total read normalization (TotalReadNormalized)log/ … log files
3.4. checkQC.py: Quality check of the input samples#
Quality check (QC) is an important step in verifying the reliability of the results obtained.
From verion 0.11.0. Churros provides a script checkQC.py to check the quality of all input samples.
build=hg38
checkQC.py Churros_result/$build/churros.QCstats.tsv samplepairlist.txt
If the samples do not meet the criteria, the script will output a warning message.
See the checkQC.py: check the quality of the input ChIP-seq samples page for the detailed criteria.
3.5. churros_callpeak: call peaks by MACS2#
churros_callpeak calls peaks of the samples specified in samplepairlist.txt using MACS2.
If input samples are omitted, peaks are called using ChIP samples only.
churros_callpeak -p 8 samplepairlist.txt hg38
churros_callpeak also outputs the correlation scores (Simpson index) and heatmaps.
- Output
macs/ … peak files called by MACS2. The log files are stored in *log.
samplepairlist.txtinmacs/directory includes the filename of peak files that is used inchurros_visualize.
3.6. churros_visualize: visualize read distributions by DROMPA+#
churros_visualize visualizes the distribution of raw reads, ChIP/Input enrichment and ChIP/Input p-value in PDF format.
The pdf files and corresponding peak lists are generated in pdf/.
churros_visualize samplepairlist.txt drompa+ hg38 Referencedata_hg38
To specify binsize 5-kbp, supply -b 5000. -l 8000 means the line size for each page is 8-Mbp. -P "--scale_tag 100" indicates the scale of y-axis is 100.
churros_visualize -b 5000 -l 8000 -P "--scale_tag 100" samplepairlist.txt \
drompa+.bin5M hg38 Referencedata_hg38
3.6.1. Highlight peak regions#
churros_visualize can highlight peak regions if the peak file is specified in samplepairlist.txt.samplepairlist.txt for churros_visualize is <ChIP-sample>,<Input-sample>,<prefix>,<peakfile>).churros_callpeak generated Churros_result/$build/macs/samplepairlist.txt that includes the peak files, churros_visualize highlights the peak regions by the command below:samplepairlist=Churros_result/hg38/macs/samplepairlist.txt
churros_visualize $samplepairlist drompa+.macspeak hg38 Referencedata_hg38
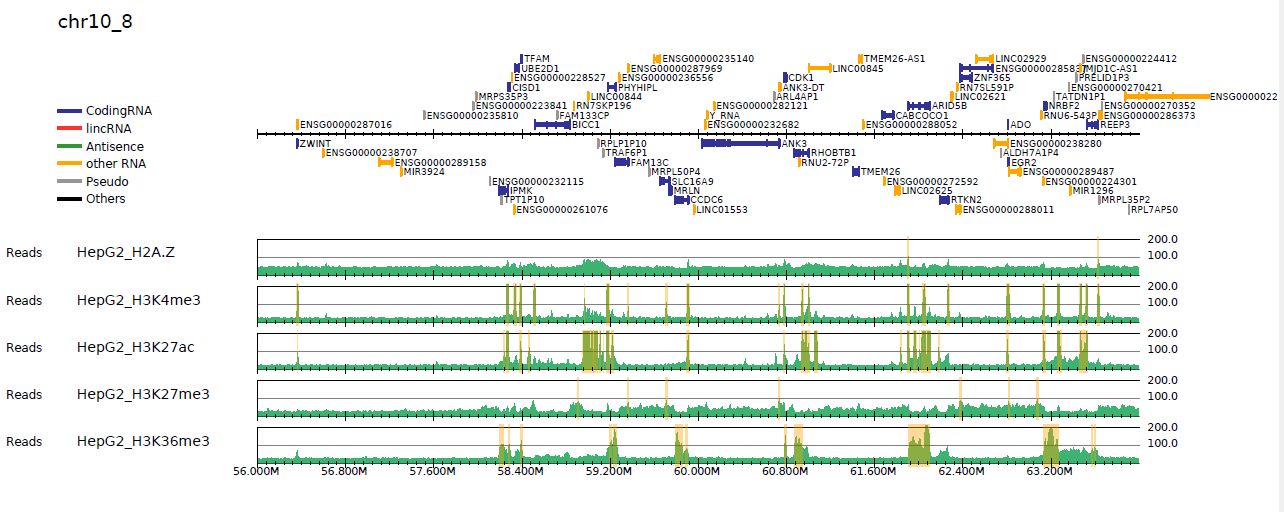
Fig. 3.1 Read distribution with peak highlighting#
3.6.2. Visualize p-value distribution#
Supply --pvalue option to visualize -log10(p) distribution of ChIP/input enrichment, which is recommended by ROADMAP project to distinguish the signal from the noise.
churros_visualize --pvalue -b 5000 -l 8000 \
samplepairlist.txt drompa+.pval.bin5M hg38 Referencedata_hg38
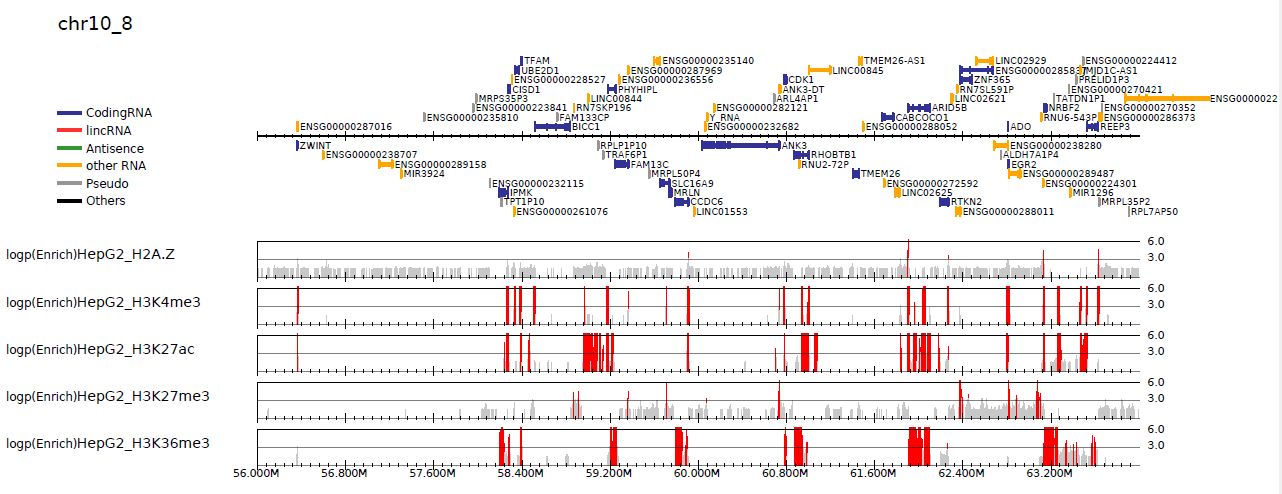
Fig. 3.2 -log10(p) distribution (ChIP/Input)#
3.6.3. Chromosome-wide view#
To visualize genome-wide view, supply -G option.
churros_visualize -G samplepairlist.txt drompa+ hg38 Referencedata_hg38
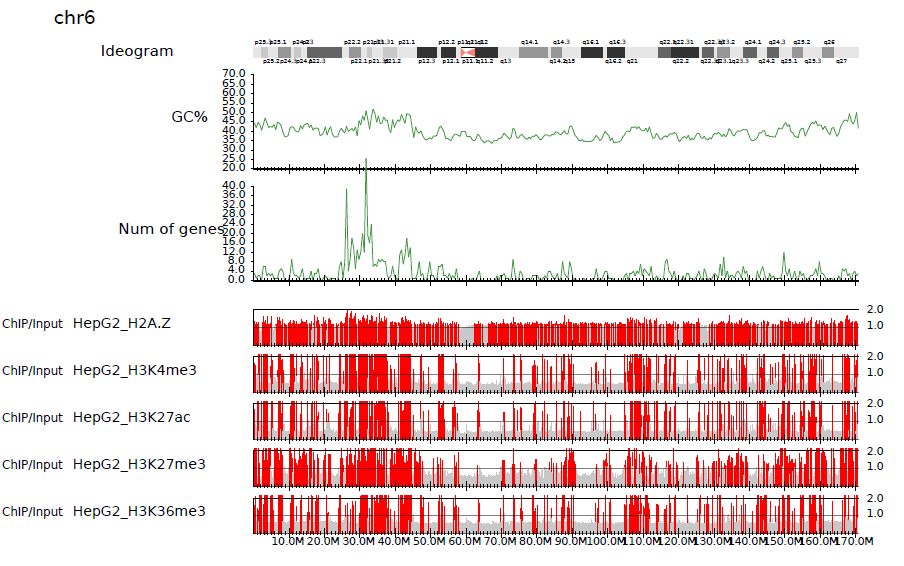
Fig. 3.3 Chromosome-wide distribution (ChIP/Input enrichment)#
3.6.4. Modify parameter sets for visualization manually#
churros_visualize also outputs a log file of pdf files generation
(e.g., `Churros_result/$build/log/pdf/drompa+.PCSHARP.100.log for Churros_result/$build/pdf/drompa+.PCSHARP.100.*.pdf).
This log file contains the command of DROMPA+ to make the pdf file at the top.
head -n1 Churros_result/$build/log/pdf/drompa+.PCSHARP.100.log
The output will look like this:
drompa+ PC_SHARP --ls 1000 -g Referencedata_hg38/gtf_chrUCSC/chr.gene.refFlat \
--gt Referencedata_hg38/genometable.txt --callpeak --showchr \
-i Churros_result/parse2wigdir+/HepG2_H2A.Z-bowtie2-hg38-raw-mpbl-GR.100.bw,Churros_result/parse2wigdir+/HepG2_Control-bowtie2-hg38-raw-mpbl-GR.100.bw,HepG2_H2A.Z, \
-i Churros_result/parse2wigdir+/HepG2_H3K4me3-bowtie2-hg38-raw-mpbl-GR.100.bw,Churros_result/parse2wigdir+/HepG2_Control-bowtie2-hg38-raw-mpbl-GR.100.bw,HepG2_H3K4me3, \
-i Churros_result/parse2wigdir+/HepG2_H3K27ac-bowtie2-hg38-raw-mpbl-GR.100.bw,Churros_result/parse2wigdir+/HepG2_Control-bowtie2-hg38-raw-mpbl-GR.100.bw,HepG2_H3K27ac, \
-i Churros_result/parse2wigdir+/HepG2_H3K27me3-bowtie2-hg38-raw-mpbl-GR.100.bw,Churros_result/parse2wigdir+/HepG2_Control-bowtie2-hg38-raw-mpbl-GR.100.bw,HepG2_H3K27me3, \
-i Churros_result/parse2wigdir+/HepG2_H3K36me3-bowtie2-hg38-raw-mpbl-GR.100.bw,Churros_result/parse2wigdir+/HepG2_Control-bowtie2-hg38-raw-mpbl-GR.100.bw,HepG2_H3K36me3, \
-o Churros_result/pdf/drompa+.PCSHARP.100 \
| tee -a Churros_result/pdf/drompa+.PCSHARP.100.log
Therefore, you can modify the resulting pdf files by directly modifying this command and -o option that specifies the output name.
For example, if you want to change the y-axis scale to 50, add --scale_tag 50 and execute:
drompa+ PC_SHARP --scale_tag 50 --ls 1000 (...) \
-o Churros_result/pdf/drompa+.PCSHARP.100.modified
See DROMPAplus manual for the detailed usage of DROMPA+.
3.7. churros_compare: compare peaks among ChIP samples#
churros_compare outputs the heatmap of the correlation of peaks between ChIP samples.
The results are output to the comparsion/ directory.
Note
By default, the churros command does not include this step because the computation time becomes long when the number of samples is quite large. Add the --comparative option to include this step in churros.
It the number of peaks largely varies among samples, the comparison may become unfair. Therefore churros_compare also estimates peak overlap for ‘top-ranked 2000 peaks’.
churros_compare samplelist.txt samplepairlist.txt hg38
- The results include three types of comparisons.
bigwigCorrelation/… Spearman correlation of read distributions in 100 bp and 100 kbp bins from deepTools plotCorrelation. This score evaluates the similarity of the entire genome including non-peak regions. Therefore, the results may reflect the genome-wide features (e.g., GC bias and copy number variation) rather than peak overlap.Peak_BPlevel_overlap/… results of the base-pair level overlap of peaks (Jaccard index) using BEDtools jaccard. This score is good for broad peaks such as some histone modifications (H3K27me3 and H3K36me3).Peak_Number_overlap/… results of peak-number level comparison (Simpson index).PairwiseComparison/contains the results of all pairs (overlapped peak list and Venn diagram) and thePeakscontains the top-ranked peaks of samples. This score is good for comparing sharp peaks such as transcription factors.
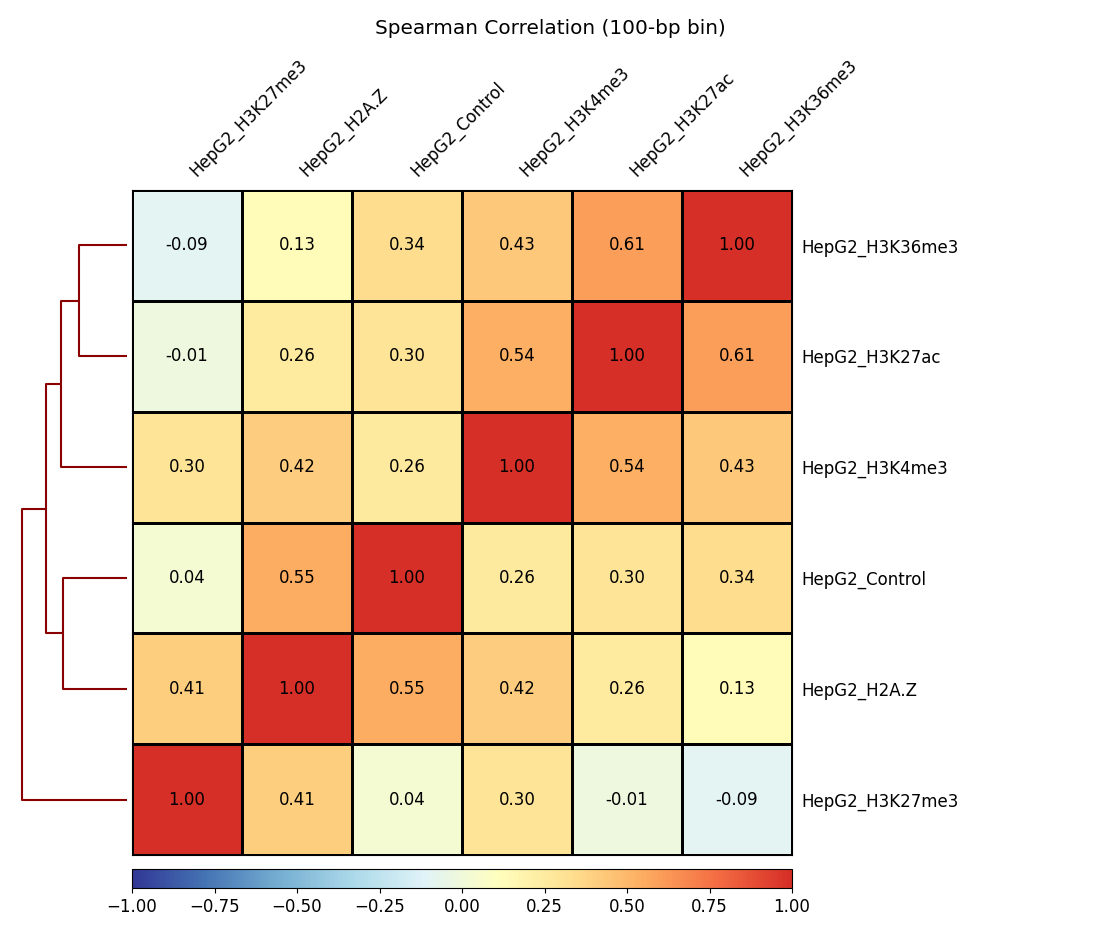
Fig. 3.4 bigwigCorrelation#
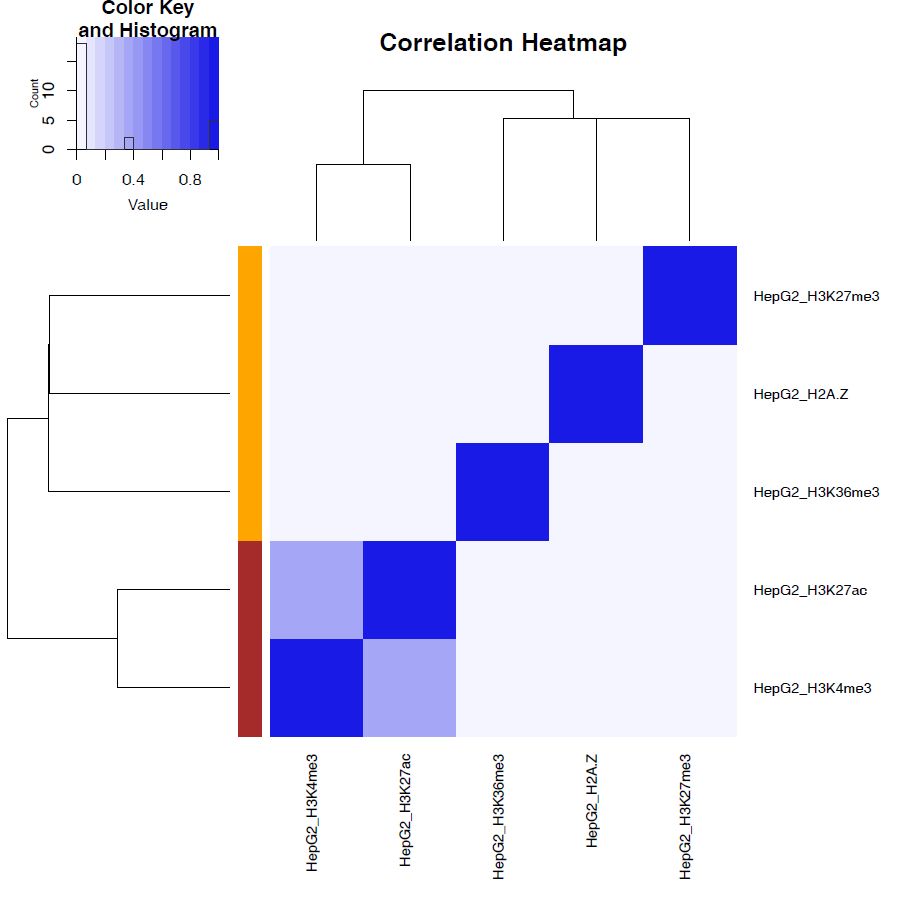
Fig. 3.5 Peak_Number_overlap#
3.8. churros_genPvalwig: generate P-value distribution as bedGraph#
churros_genPvalwig generates a -log10(P-value) distribution in bedGraph format. The P-value of upregulation and downregulation is output separately. This bedGraph file is suitable for the ChIP-seq imputation. The results are output in drompa+.pval/.
Note
By default, the churros command does not include this step. Add the --outputpvalue option to include this step in churros.
Ddir=Referencedata_hg38
gt=$Ddir/genometable.txt
churros_genPvalwig samplepairlist.txt drompa+.pval hg38 $gt
